WP2
SofTMech Work Package WP2 Progress Report
M. Chaplain, C. Macnamara
August 2021
2.1 Mechanobiological models of cell-cell and cell-ECM interactions
Background
In both normal and pathological settings, the tissue microenvironment influences cell functionality. How cells maintain and remodel the extracellular matrix (ECM) is vitally important both for tissue homeostasis and for pathological scenarios such as tumour growth, cancer invasion and myocardial infarction (MI). Activating invasion and metastasis is one of the key “hallmarks of cancer”. Loss of cell-cell adhesion, modification of cell-matrix adhesion (e.g. through integrin expression) and interaction with the (highly heterogeneous) tumour microenvironment are all crucial individual components of invasion and metastatic spread of cancer. In cardiac repair, the recruitment and differentiation of peripheral connective tissue cells following MI are also tightly regulated by the local environment. In this work package, we have been developing a multiscale model to investigate the complexities of the interactions of the various components.
Work done so far
We have developed a computational code which is being used as the basis to simulate the force-based, individual-based model of cell-cell, cell-matrix and cell-blood-vessel interactions. The model was implemented to model the growth of cancer cells around a central blood vessel – the “tumour cord” or “tumour cuff”. The new code was extended to include individual fibres in 3-dimensions and thus enable the explicit modelling of cell-matrix interactions. Work on this Work Package was effectively stopped from October 2017 until January 2019 due to the fact that the PDRA working on this WP, Dr Cicely Macnamara, was on maternity leave. Work recommenced between January 2019 and October 2019, but stopped again due to a second period of maternity leave for Dr Macnamara. The second period of maternity leave was from 1 October 2019 – 29 September 2020. However, on returning, work was again slowed down due to COVID-19 restrictions and home schooling.
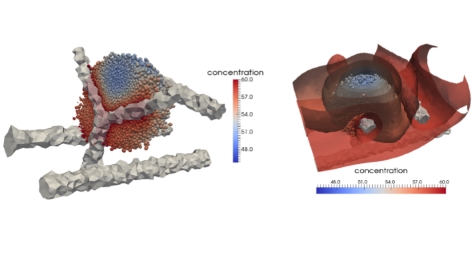
Nonetheless we have published a paper detailing the main results of the model in the Journal of Computational Science. The work focused on understanding the specific mechanisms that occur in the tumour microenvironment and we have developed a 3D individual-based, force-based model which allows one to simulate the behaviour of, and spatio-temporal interactions between, cells, extracellular matrix fibres and blood vessels. Each agent (e.g. a single cell, a single fibre of the ECM) is fully realised within the model and interactions are primarily governed by mechanical forces between elements. As well as the mechanical interactions we have also developed the model to consider chemical interactions, by coupling the individual-based code to a finite element solver to model the diffusion of oxygen from blood vessels to cells. The current state of the art of the model allows us to simulate tumour growth around an arbitrary blood-vessel network or along the striations of fibrous tissue. Other recent work has involved (i) the development of a mathematical framework for the metastatic spread of cancer through local invasion of the ECM (interactions with the tumour microenvironment), interactions with blood vessels (intravasation), transport through the blood vessels/circulatory system, escape through vessel walls (extravasation) and then re-growth at the secondary location. This is the first explicitly spatial model of metastatic spread and has now been published in the Bulletin of Mathematical Biology and the IMA Journal of Applied Mathematics; (ii) the development of a model of pattern formation of cell aggregation using a development of the mechano-chemical framework of Murray & Oster. The work here has shown new insight into the role of cell-tissue constitutive equations governing the ability to form cellular patterns or not.
Interaction with WP1 (incorporating the results of the individual cell modelling) is now currently underway, as is interaction with WP3 (upscaling) once we have undertaken some more intensive computational simulations of the individual-based model. We have also started collaborating with Prof Steven McDougall (WP6) to integrate the 3D individual-based, force-based model with Prof McDougall’s angiogenesis model. The aim is to focus attention on intravasation of cancer cells into blood vessels (microvasculature), and then the transport of the cancer cells into the main blood system.
Output:
Franssen, L., Lorenzi, T., Burgess, A.E.F., Chaplain, M.A.J. (2019) A mathematical framework for modelling the metastatic spread of cancer. Bull. Math. Biol. 81, 1965-2010. 10.1007/s11538-019-00597-x
Macnamara, CK, Caiazzo, A, Ramis-Conde, I, Chaplain MAJ (2020) Computational modelling and simulation of cancer growth and migration within a 3D heterogeneous tissue: The effects of fibre and vascular structure. J. Comput. Sci. 40, 101067
Prof M Chaplain – Plenary Speaker, 6th International Conference on Computational and Mathematical Biomedical Engineering, CMBE 2019, Tohoku University, Katahira Campus, Sendai, Japan, 10-12 June 2019.
Franssen, L.C., Sfakianakis, N., Chaplain, M.A.J. (2021) "A novel 3D atomistic-continuum cancer invasion model: In silico simulations of an in vitro organotypic invasion assay" J. Theor. Biol. In press, doi:10.1016/j.jtbi.2021.110677.
Macnamara, C.K. (2021) "Biomechanical modelling of cancer: Agent-based force-based models of solid tumours within the context of the tumour microenvironment" Comp. Sys. Onco. 1(2), https://doi.org/10.1002/cso2.1018
......................................................................................................................
Mechanobiological Models of Cell-Cell and Cell-ECM interactions (update:May 2018)
The tissue microenvironment influences cell functionality. How cells maintain and remodel the extracellular matrix (ECM) is vitally important both for tissue homeostasis and for pathological scenarios such as tumour growth, cancer invasion and myocardial infarction (MI).
Project 1: We will adopt an individual-based, force-based modelling approach to investigate cell-cell, cell-fibre and cell-ECM interactions using our multiscale-multicompartment model.
Project2: By taking into account the results of WP1 concerning the biomechanical properties of the cells and including the effect of actin and integrins on cell-ECM adhesion, we will develop a new system of intracellular ODEs governing these dynamics.
Project3: model extension through computational upscalling
Project4: Development of a fully 3D multiscale computational model of a growing, invading cell mass, which will be applied to the specific cases of breast cancer through WP6
Team: Prof. Chaplain (Team leader), Dr. McDougall, Prof. Ogden, Dr. Yin, Prof. Olson, Prof. Insall,
Prof. Husmeier, Prof. Luo, PDRA2, PhD2, PhD3
Progress
We have developed a 3D computational code which is being used as the basis to simulate the force-based, individual-based model of cell-cell, cell-matrix and cell-blood-vessel interactions.
The model has now been implemented to model the growth of cancer cells around a central blood vessel - the “tumour cord” or “tumour cuff”. In this scenario, tumour cells grow around a central blood vessel with those cells further away from the blood vessel experiencing lower nutrient levels. The work carried out here was the first to adopt an individual-based model to examine the growth of tumour cords and was able to estimate the distance from the blood vessel that cancer cells first became necrotic.
The new code has also been extended to include individual fibres in 3-dimensions and thus enable the explicit modelling of cell-matrix interactions. This will be used for the basis of a model of cancer invasion of the extracellular matrix (ECM) as well as for the first simulations of myocardial infarction. Additionally we have extended the code to solve numerically PDEs in 3D and thus provide the computational solution to external chemical fields, such as oxygen. This has been done in such a way as to be compatible and integrable with the software being developed in WP6 by Professor McDougall and the initial modelling of perfusion already carried out there. In parallel, we have developed computational method and experimental studies on cell migration and chemotaxis.